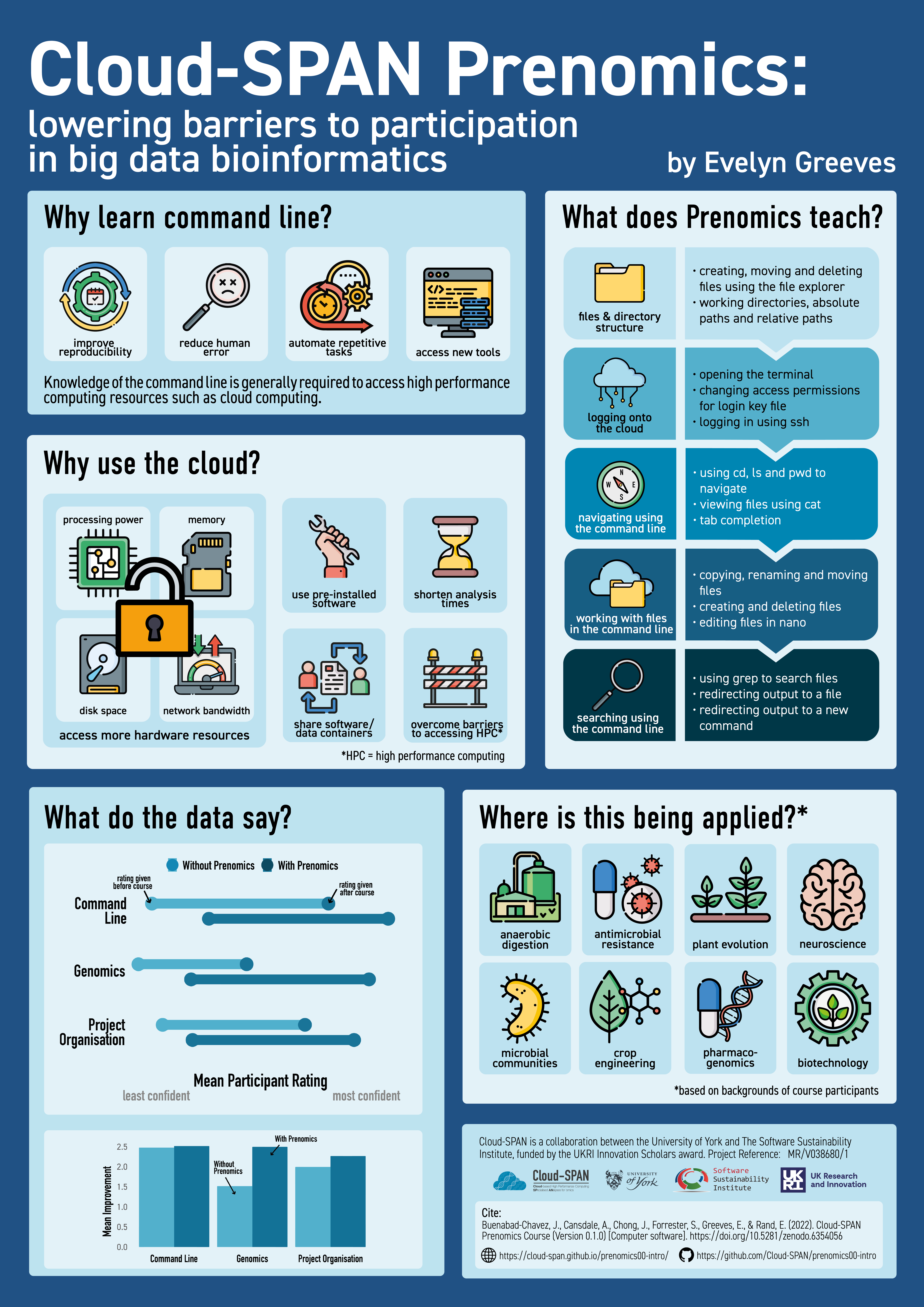
I designed this poster for the University of York Biology Department’s Research Away Day in September 2022. The aim was to demonstrate how providing learners with more support at the early stages of learning command line programming can help them further down the line.
When we ran our first Genomics course in autumn 2021, we discovered that a major sticking point for many participants was directory structure and navigation. This was making it really difficult to help people log into the cloud, where we could then teach them core command line skills. We spent much more time troubleshooting and going over these basic skills than we’d expected, meaning we had to rush through more complex content later on about analysis tools and pipelines.
Overall, this meant our learners began their command line journey from a place of frustration (which has a huge impact on confidence) and we were forced to minimise the amount of time spent on the ‘genomics’ part of the Genomics course.
As a result, we split the original Genomics curriculum across two courses: Prenomics and Genomics. In Prenomics, all the focus is on the command line. We teach about files and directory structure, and spend lots of time guiding learners through creating a folder and navigating to it without using the command line. We also teach those crucial core skills - commonly used commands like ls, cd and mv. Later, as learners gain confidence, we move on to more complex commands like grep and redirection with > and >>.
The poster itself is based on a previous poster I made about contents of Prenomics, but with an added section demonstrating that learners ended Genomics with a higher level of understanding across all three topic areas when Prenomics was offered as a separate course. They also saw a greater mean improvement in understanding when Prenomics was offered, especially for the Genomics topic - likely because more time was able to be dedicated to teaching these more complex ideas.
Making the poster
I always enjoy graphic design and this was a fun challenge because it could be very bold and graphic, with lots of sections to fit together in a kind of jigsaw. I designed the icons in Adobe Illustrator and put the poster together in Adobe InDesign. The graphs at the bottom were made mostly in RStudio using ggplot2, with the axis labels and annotations added afterwards in Illustrator to get the fonts to match the rest of the poster (although I now know how to do this in ggplot!).
The finished product
(click to enlarge)